Research background
In nature, NAD(P) -dependent oxidoreductases are one of the most widely distributed enzymes, and they play an important role in the energy metabolism and REDOX reaction of organisms.
NAD(P) acts as a cofactor for these enzymes, and its specificity (that is, the enzyme’s selectivity for NAD or NADP) is critical for the enzyme’s function and activity.
Identifying cofactor preferences for these enzymes on a large scale and designing mutants to switch cofactor specificity remain complex tasks.
Traditional experimental methods are not only time-consuming and labor-intensive, but also inefficient, which can not meet the needs of high-throughput protein engineering.
It is important to develop a method that can accurately predict and engineer the specificity of NAD/NADP cofactors.
With the rapid development of deep learning technology, its application in the field of bioinformatics is increasingly extensive.
The Transformer model, as an attention-mechanism-based deep learning architecture, is excellent at processing sequence data and capturing long-distance dependencies.
This makes the Transformer model a powerful tool for solving protein sequence analysis and design problems.
Especially in terms of enzyme cofactor prediction, Transformer model can predict enzyme selectivity for NAD or NADP by learning characteristic patterns in protein sequences.
Research questions:
- How can we use deep learning techniques, especially the Transformer model, to accurately predict the cofactor specificity of NAD/ NADP-dependent REDOX enzymes?
- How can key residues affecting cofactor specificity be revealed by analyzing the attention layer of the Transformer model?
- How can these findings be applied to enzyme engineering to design mutants with desired cofactor specificity?
Research result
1. Construction and validation of the DISCODE model
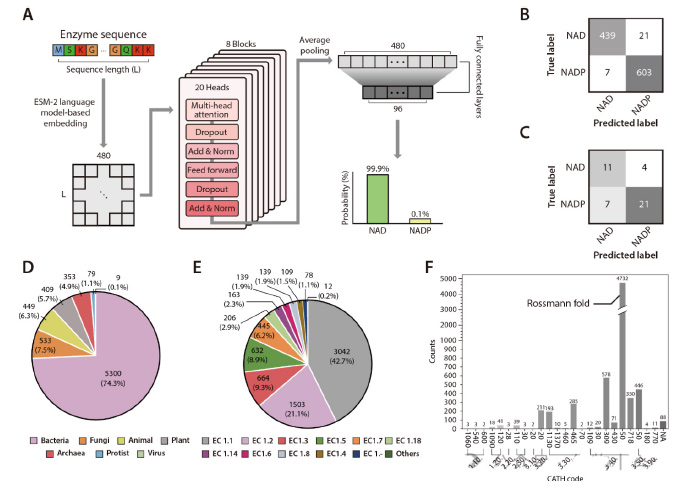
The research team developed a Transformer model called DISCODE, a deep learn-based iterative pipeline for analyzing the specificity of cofactors and designing enzymes.
This model can use the sequence information of NAD and NADP binding proteins to predict the cofactor specificity of enzymes.
By benchmarking against existing cofactor prediction models such as Cofactory and Rossmann-toolbox, the DISCODE model performed well in both prediction accuracy and F1 scores.
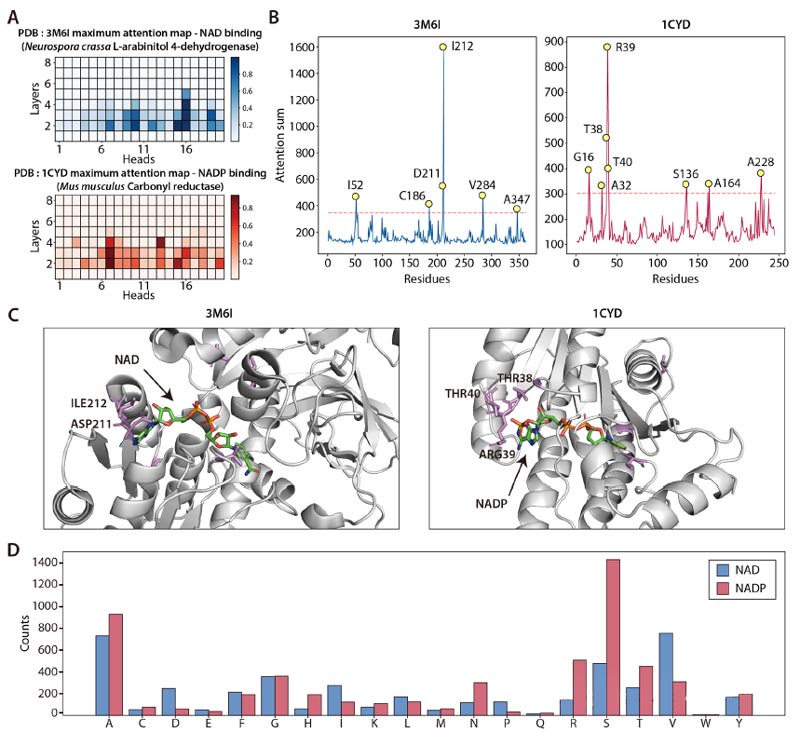
2. Attention Layer analysis reveals key residues
By analyzing the attention layer of the DISCODE model, the research team found a number of residues that showed significantly higher attention weights when predicting cofactor specificity.
These residues are closely related to structurally important residues that interact closely with NAD(P), thereby identifying key residues that affect cofactor specificity.
These key residues were highly consistent with verified cofactor switching mutants, indicating the effectiveness of attention-level analysis in identifying key residues.

3. Use attention analysis to guide enzyme engineering
The research team combined the DISCODE model with attention analysis to build a fully automated enzyme design pipeline.
The pipeline is able to identify potential replacement sites, generate mutant sequences, and assess the cofactor preferences of each mutant.
With this approach, the research team successfully engineered mutants with the desired cofactor specificity.
For example, when analyzing Neurospora crassa L-Arabitol 4-dehydrogenase (PDB:3M6I), attention analysis identified key residues such as T38, R39, and T40.
These residues are located in the structural modes and are critical for interacting with the 2′ -phosphate group of NADP.
By mutating these residues, the team managed to switch cofactor specific.
4. Validation experiment and model optimization
To verify the prediction accuracy of the DISCODE model, the research team conducted extensive experimental validation.
They selected multiple NAD/NADP dependent REDOX enzymes and constructed corresponding mutants.
By comparing the cofactor preferences of wild-type and mutants, the team found that the predictions of the DISCODE model were highly consistent with the experimental results.
The research team also optimized the DISCODE model based on the experimental results to improve its predictive performance.
Research significance
Significant progress has been made in NAD/NADP cofactor prediction and engineering by developing DISCODE models and utilizing Transformer attention analysis technology.
These findings not only promote the development of related fields,
but also provide new ideas and methods for other bioinformatics problems.
Literature source:
Kim J, Woo J, Park JY, Kim KJ, Kim D. Deep learning for NAD/NADP cofactor prediction and engineering using transformer attention analysis in enzymes. Metab Eng. 2025 Jan;87:86-94. doi: 10.1016/j.ymben.2024.11.007. Epub 2024 Nov 20. PMID: 39571721.